I was excited to see a preprint article of a study on genetic factors that influence SARS-CoV2 susceptibility (published on May 7th). The study was performed by scientists at the University of California, San Francisco. You can read through the whole study here: The landscape of host genetic factors involved in infection to common viruses and SARS-CoV-2
The researchers used genetic sequencing data from the UK Biobank, which anonymously shares the genetic data of people in the UK who have opted into the research. The UK Biobank had recently announced that they had genetic information on individuals who had tested positive for COVID-19.
For this study, the researchers used genetic data plus information on viral infections for a number of different viruses, including this current SARS-CoV2 virus. The purpose of the study was to highlight genes that were related to genetic susceptibility to various viral pathogens.
Genetic variants linked to COVID-19 infections
Cutting to the chase here, the researchers linked a number of different genetic variants to the susceptibility to SARS-CoV2.
Before I list the genes, let me explain a bit about what the statistics mean.
First, a genome-wide study (GWAS) is going to show the more common variants linked to a phenotype. This type of study usually won’t include mutations that may be important but are only found in a small percentage of the population. Recent serology studies are showing that most people who have the virus are asymptomatic. If 94% (NY numbers announce by Cuomo) to 99% of people are asymptomatic[ref], rare genetic variants may be really important here, especially when it comes to younger people who have COVID-19 symptoms.
The research lists the ORs (odds ratios) for the variants.
- An odds ratio of 1 (OR=1) means that there is neither an increase nor a decrease in risk.
- If the odds ratio is greater than 1, the relative risk of something increases. For example, if the OR=1.35, there is a 35% increase in the relative risk of something happening. In this case, if your risk of getting COVID-19 is 6 in 100, then a 35% increase in risk would mean that your risk is 8.1 in 100.
- If the odds ratio listed is less than 1, then the relative risk of something decreases — you are less likely to have that condition occur. For example, an OR=0.7 means that you are at a 30% decreased relative risk of something happening.
Finally, I also want to point out that only 1,028 individuals were included in the genome-wide association analysis. That is a much smaller sample size than is usually included in GWAS studies due to only having the genetic data for that number of SARS-CoV2 patients.
More genetic studies will come out on this topic. And different genetic variants are likely to be important. So please don’t take this as the ‘final word’ on this topic. Generally, the variants that pop up in a genome-wide association study (GWAS) point to the genetic pathways that are important, rather than specific SNPs.
One last note: this study was done on UK biobank participants (mostly Caucasian population). It is likely that different genetic variants will be important for different population groups.
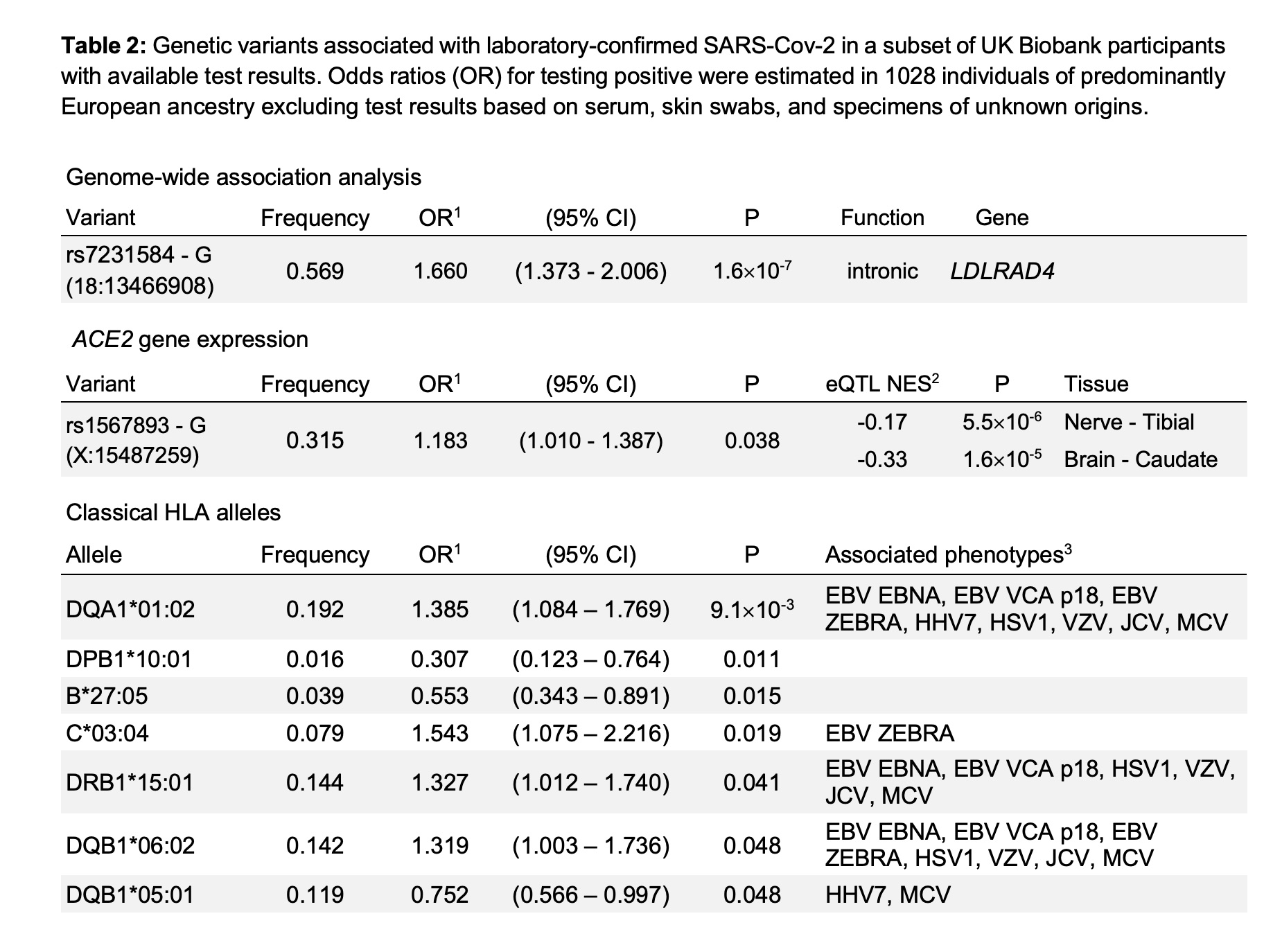
Here is the listing of variants impacting the risk of SARS-CoV2 infection from the preprint that was published (creative commons license):[ref]
Does the LDL cholesterol receptor variant impact COVID-19 risk?
The first variant listed, rs7231584, is in the LDLRAD4 gene. This is an LDL cholesterol receptor gene, also known as C18orf1.[ref]
One connection between LDLRAD4 and viral infections shows LDLRAD4 impacting the pro-inflammatory cytokine, TGF-β. TGF-beta also may be important in fibrosis in the lung tissue.[ref][ref]
Other studies on COVID-19 patients noted that low LDL cholesterol levels are linked with higher mortality rates.[ref] This is not unique to coronavirus infections. Low LDL cholesterol levels are a risk factor for many pathogens. Pathogens need an optimal level of cholesterol in the cell wall for infection.[ref] Cholesterol is also important in innate immunity.[ref]
The rs7231584 variant is not found in 23andMe or AncestryDNA data, nor are there any other studies on that variant. Carrying the more common allele for this variant increased the relative risk of COVID-19 by 60%. (Another way of looking at it is that the less common variant protected against infection.)
ACE2 variant shows low relative risk:
The second variant linked to SARS-CoV2 susceptibility is in the ACE2 gene. This gene codes for the angiotensin-converting enzyme 2, which is the cell receptor through which the coronavirus enters the host cells.
The variant – rs1567893 – is not available in 23andMe, AncestryDNA, etc data sets and there are no other studies on this variant. The minor allele here increases the relative risk of infection by 18% (OR = 1.18), which is not much of an impact.
HLA types and their role within the immune system:
It is not at all surprising that different HLA types are nominally associated with increased or decreased risk of COVID-19. We all carry different HLA types — and it is through all of this variation that we survive as a human species!
HLA stands for human leukocyte antigen, and it is an important – and variable – part of the immune system. These are the responsible proteins that let the immune system know what is going on in a cell. On the surface of nearly every cell in the body, the HLA proteins can present peptides from inside the cell to be detected by the immune system. If the immune system recognized the peptide as ‘foreign’, it then will attack the cell, causing it to self-destruct.
The HLA system is comprised of more than 200 different genes, which are classified into two main classes, with subclasses within. All of these have genetic variants, thus giving rise to the ability for some people to be protected against different pathogens. Even novel pathogens, like this new coronavirus, won’t wipe out everyone because we humans have such different innate immune systems.[ref]
Diversity and variability allow us to thrive as a species.
The HLA system has typically been determined using serology testing – like testing for your A, B, O blood type. Some of the HLA variants are mapped to specific genetic variants, but others are tagged by combos of variants — or are unknown at this point.
Genetics of HLA types:
The SNPs listed below are linked to — but not 100% definitive for – the HLA types.
Members: Log in to see your data file Not a member? Join now.
HLA-DQA1*01:02
It is also associated with either increased or decreased susceptibility to other viruses, including Epstein-Barr. The Broad institute lists the SNP tags for HLA-DQA1*01:02 as rs8180664 or rs3134975.[ref] Others list rs9269910 as a tagging SNP.
Check your genetic data for rs8180664 (23andMe v4, v5):
- T/T: likely a carrier for HLA-DQA1*01:02, possibly slightly increased risk of SARS-CoV2 infection (OR=1.3)
- C/T: likely a carrier of HLA-DQA1*01:02
- C/C: typical
Members: Your genotype for rs8180664 is —.
HLA-B27 allele:
Related article: HLA-B27 and autoimmune diseases
Check your genetic data for rs4349859 (23andMe v5 only; AncestryDNA):
- A/A: high likelihood of carrying 2 copies of HLA-B27[ref][ref][ref][ref] decreased risk of SARS-CoV2
- A/G: high likelihood of carrying 1 copy of HLA-B27; decreased risk of SARS-CoV2
- G/G: typical
Members: Your genotype for rs4349859 is —.
HLA-DRB1*1501
Check your genetic data for rs3135388 (23andMe v4, v5; AncestryDNA):
- G/G: typical
- A/G: possibly increased risk of SARS-CoV2 infection (OR=1.3)
- A/A: possibly increased risk of SARS-CoV2 infection (OR=1.3)[ref]
Members: Your genotype for rs3135388 is —.
HLA-DQB1*06:02 is not available in 23andMe data (that I know of). Interestingly, this HLA type is highly linked to narcolepsy susceptibility.[ref]
Taking all of this with a grain of salt…
The researchers conducting the study conclude with: “While the reported associations for SARS-CoV-2 were based on a limited sample size, we believe these findings may be informative when considered in the context of our results for other viral infections, and contribute to the global, pressing need to understand COVID-19, which motivated our rapid analysis of these data. However, care must be taken with the interpretation of genetic associations with SARS-CoV-2 status, which should be regarded as preliminary and exploratory given the small number of tests within a biased sample, confounding by factors related to symptom recognition, access to testing, and limited statistical power. ”
Will the HLA types of individuals be important? Undoubtedly.
Will other genes related to immune response also be important? Most likely.
The biggest risk factors of severe COVID-19, though, remain as being in poor health (kidney disease, diabetes, heart disease, etc) and being elderly (less initial immune response).
Related Articles and Genes:
Acute Respiratory Distress Syndrome (ARDS) genes
ARDS is caused by overwhelming immune response to a virus, bacteria, or lung injury. Learn more about which of your immune system genes are involved in ARDS.
Viral Immunity: Your genes protect you
Your genetic variants shape your immune system and give you superpowers against some pathogens – and perhaps more susceptible to others.
Non-secretors: Norovirus resistance and gut microbiome
A genetic variant in the FUT2 gene controls whether or not you secrete your blood type into your saliva and other bodily fluids, such as your intestinal mucosa. This alters the gut microbiome – and protects you from Norovirus.